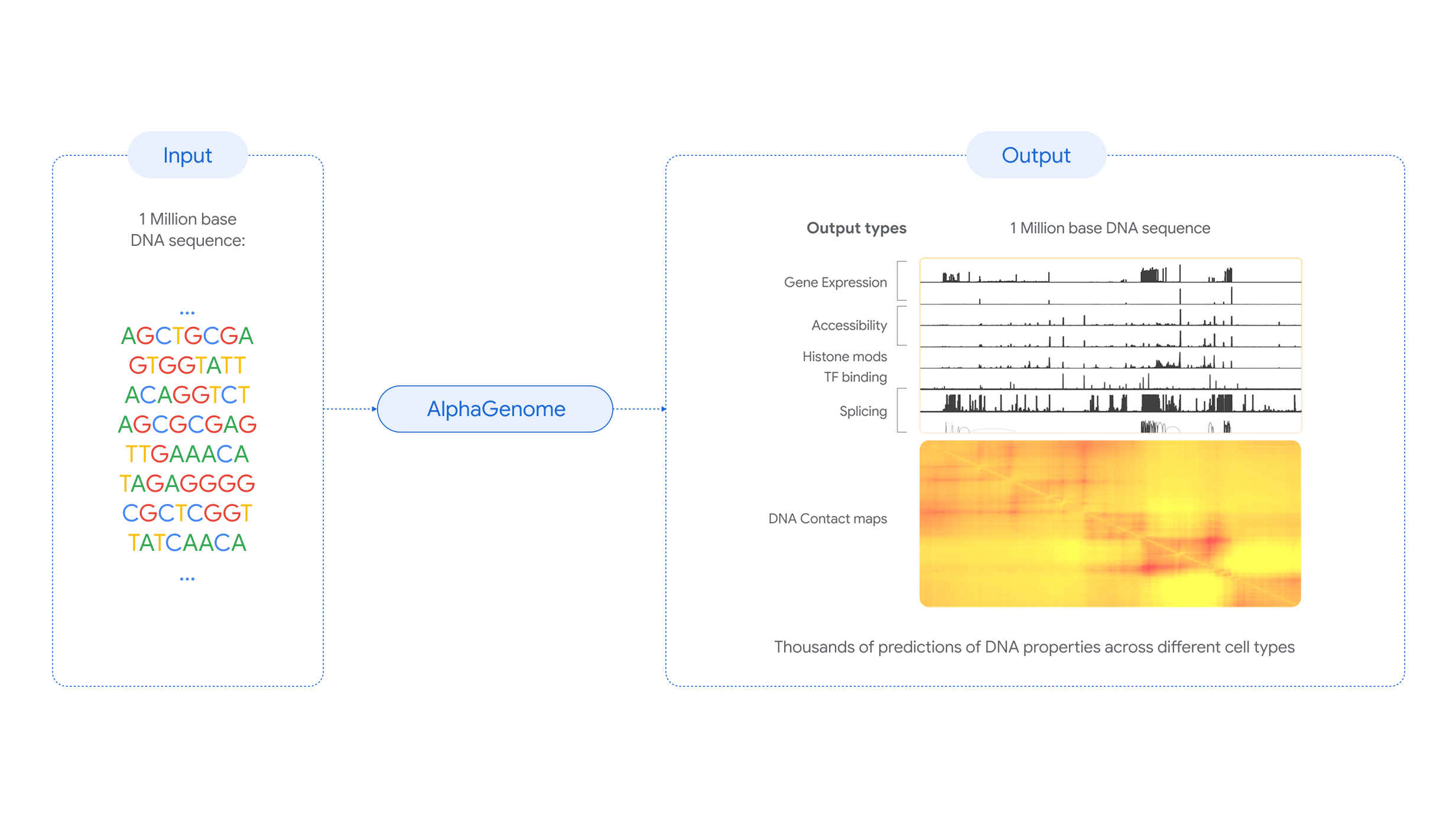
AlphaGenome, an AI tool from Google, predicts the impact of variations in DNA
AlphaGenome is a deep learning model developed by Google DeepMind capable of predicting the function of DNA sequences up to one million base pairs long. An evaluation of the tool shows that it matches or improves upon the predictive ability of existing models in 25 of the 26 tests performed. According to the authors, who are part of Google DeepMind itself, AlphaGenome can help scientists "better understand genome function, the biology of diseases, and ultimately drive new biological discoveries and the development of new treatments." The results are published in Nature.
Montoliu- AlphaGenome
Lluís Montoliu
Research professor at the National Biotechnology Centre (CNB-CSIC) and at the CIBERER-ISCIII
A mouse, a turbot, and a human have roughly the same number of genes, around 20,000 genes each. However, everyone understands that we are very different from a mouse and a turbot. Therefore, in a way, the number of genes we have doesn't tell us about our complexity or what we will be like. The solution to this paradox lies in the non-coding genome, the part that doesn't contain protein-coding information.
Remember that, in all these animals, and particularly in us, those 20,000 genes barely make up 2% of our entire genome. This surprised geneticists for many years. What's in the remaining 98% of the genome? (That is, in most of the genome!). The answer is that this dark and unknown part of the genome harbors many repetitive sequences, many mobile elements (transposons and retrotransposons), and, most importantly, the regulatory DNA sequences (to which specific proteins bind) that tell a gene when and where to start functioning, or when and where to turn off. In other words, the morphological complexity and diversity we see in a mouse, a turbot, and a human is not achieved by increasing or decreasing the number of genes, but rather by altering precisely these regulatory sequences. These sequences cause the same genes to be activated or deactivated at different times during development or in different cells, depending on the species. Essentially, if we change the activation and deactivation program of the same genes, we will generate very different animals. And that is what ultimately produces animals as disparate as these three, even though they have a similar number of genes.
For years, algorithms and computer programs have been developed to analyze this non-coding, non-meaningful part of the genome, searching for precise sequences known from other research to be regulatory elements. These elements come in many forms: some are associated with gene activation, others with gene silencing, and some even act as insulators, separating what happens in one gene from what will happen in a neighboring gene. These predictive programs are based on the systematic comparison of sequences known to operate as activators, silencers, or insulators (among many other types of regulatory elements) in a specific DNA sequence, which is usually not too long, typically a few thousand nucleotides. Each of these programs usually specializes in detecting one of these types of regulatory elements.
All of this has been turned upside down with the emergence of AlphaGenome, whose public launch has just been published in the journal Nature. A new artificial intelligence from Google, yet another one, is capable of performing all these inspections and deductions on virtually all types of regulatory elements, all at once, and it can do so on enormous segments of DNA, up to a million letters long—something we didn't know how to do.
Google already surprised the scientific community a few years ago with AlphaFold and its impressive ability to predict the structure and folding of proteins from just the DNA sequence. Now it leaves us speechless again with AlphaGenome and its ability to interpret and predict all those non-coding sequences in the genome, the non-meaning sequences found in a huge segment of our genome. To develop this AI, researchers trained it by analyzing the human and mouse genomes.
Naturally, this will have a significant impact not only on basic research, to understand how genes work, but also on more practical, applied aspects. For example, how to identify new DNA sequences that are relevant in those areas of the genome that are usually overlooked or not taken into account. The information provided by AlphaGenome must be carefully considered and analyzed when addressing genetic diagnoses.
An alteration in any of these regulatory elements, preventing the activation or silencing of a gene when it should occur, can result in a change in the pattern of embryonic development or the appearance of symptoms of a pathology associated precisely with the abnormal functioning of that gene (without the gene's sequence itself having changed at all). In this case, the mutation would not be within the gene but outside, in more or less distant sequences, in the DNA sequences that conceal these regulatory elements. This AI, called AlphaGenome, will now efficiently discover these elements in any segment of the genome we want to analyze, especially in the non-coding genome—the genome we used to call meaningless (when we didn't understand it)—which, thanks to AlphaGenome, we can now begin to decipher with much greater precision and detail than we previously knew.
Goldstone - AlphaGenome
Robert Goldstone
Head of Genomics at the Francis Crick Institute (UK)
DeepMind’s AlphaGenome represents a major milestone in the field of genomic AI. This level of resolution, particularly for non-coding DNA, is a breakthrough that moves the technology from theoretical interest to practical utility, allowing scientists to programmatically study and simulate the genetic roots of complex disease.
The model performs exceptionally well on tasks that might be expected to be governed by rigid ‘grammatical’ rules written in the DNA, such as splice site prediction. In these areas, it is poised to immediately replace older standard tools. And one of the most remarkable demonstrations is its ability to predict gene expression from DNA sequence alone. Whilst not perfect, given that gene expression is influenced by complex environmental factors that the model cannot see, achieving the level of accuracy demonstrated, based solely on ‘local' DNA rules, is an incredible technical feat.
AlphaGenome is not a magic bullet for all biological questions, but it is a foundational, high-quality tool that turns the static code of the genome into a decipherable language for discovery.
Conflicts of interest: no declarations of interest. There is a DeepMind lab at the Francis Crick Institute, but Robert has not worked on AlphaGenome.
Lehner - AlphaGenome
Ben Lehner
Head of Generative and Synthetic Genomics, Wellcome Sanger Institute (Cambridge, UK)
AlphaGenome is a great example of how AI is accelerating biological discovery and the development of therapeutics. Identifying the precise differences in our genomes that make us more or less likely to develop thousands of diseases is a key step towards developing better therapeutics. AlphaGenome and models like it that help decipher the regulatory code of our genome will make it much easier to do this.
As we have come to expect from Google Deepmind, AlphaGenome is a great piece of engineering that brings together ideas developed by many different scientists into a model that sets the standards. At the Wellcome Sanger Institute we have tested AlphaGenome using over half a million new experiments and it does indeed perform very well.
However, AlphaGenome is far from perfect and there is still a lot of work to do. AI models are only as good as the data used to train them. Most existing data in biology is not very suitable for AI - the datasets are too small and not well standardized. The most important challenge right now is how to generate the data to train the next generation of even more powerful AI models. We need to do this fast, cost effectively and in a way that both the data and the resulting models are available for everyone to use.
Conflicts of interest: Ben Lehner has some research funding from Google DeepMind / a small collaboration with them. Ben is not an author on, and was not involved in the development of, the AlphaGenome model.
Avsec et al.
- Research article
- Peer reviewed