A new genome editing technique using 'RNA bridges'
Two articles published in Nature describe a new genome editing technique that enables the insertion, inversion, or deletion of long DNA sequences at specific positions in the genome. This is a one-step approach that could offer a simpler method for genome editing in the future. The authors describe a technique to create reprogrammable recombinases—key enzymes in genetic recombination. These enzymes are guided by RNA, which acts as a bridge, directing the recombinase to target sites and facilitating predetermined editing.

Montoliu - Puente ARN EN
Lluís Montoliu
Research professor at the National Biotechnology Centre (CNB-CSIC) and at the CIBERER-ISCIII
CRISPR-Cas9 genetic editing tools have extraordinary potential, but they are neither infallible nor omnipotent. They efficiently inactivate specific genes by generating mutations, which is their main strength. However, precise substitution of nucleotides (genome letters) or the insertion, deletion, or inversion of significant DNA fragments is less reliable, with high levels of uncertainty. This led to the development of second-generation CRISPR tools (base editors) and third-generation tools (quality editors), introduced by David Liu (BROAD Institute, Boston) in 2016 and 2019, respectively. Base editors robustly substitute a nucleotide according to certain possible combinations (A→G, C→T, C→G), and quality editors enable the introduction, deletion, or inversion of small DNA sequences. Nevertheless, manipulating large genome segments to insert, delete, or invert them remains a challenge, with high levels of uncertainty.
In 2021, Marc Güell's laboratory (UPF, Barcelona) introduced a new hybrid tool, elegantly combining the nuclease domain of the Cas9 protein from a CRISPR system with a transposase (an enzyme from transposons needed for these mobile elements to cut themselves out of one site and jump to another in the genome). This new tool was named FiCAT (Find and cut-and-transfer mammalian genome engineering). The FiCAT system allowed for the transfer of sizable DNA fragments (<10 kb) with about 25-30% efficiency in mouse and human cells, although it left a mark from the jump by also inserting the terminal inverted and repeated sequences (ITRs) that flank the transposon and that this mobile element needs to mobilize the DNA fragment to be inserted.
Patrick D. Hsu's laboratory (Univ. Berkeley, California) has raised the stakes with a new editing system recently presented in two articles published in Nature. The first article describes this new way of modifying DNA through programmable recombination directed by a small RNA molecule called bridge RNA. Hsu describes these components in certain mobile IS elements (insertion sequence elements), specifically with the IS110 element, and shows that this small bridge RNA molecule contains two sequences complementary to the donor DNA and the recipient DNA, which can be varied at will to direct the insertion of DNA segments of up to ~5 kb specifically to a determined location in the genome. Similarly, the same IS element system allows for the excision (deletion) or inversion of large DNA segments, and this time it does so without leaving any trace or scar at the insertion site, unlike the FiCAT system. This is surprising and very promising, with high insertion efficiencies (~85% efficiency) but also considerable insertions at unexpected genome sites, similar but not exact, in up to ~30% of cases. Comparable results are obtained with deletions and inversions, with an effectiveness range varying between ~32% and ~99% for deletions and between ~5% and ~98% for inversions.
The most surprising aspect of this new tool, which still needs further optimization and improvement, is its high level of compactness, with a bridge RNA of just 150-250 nucleotides and a small recombinase enzyme encoded by just 300-460 amino acids that fit within this IS110 mobile element. The results presented have only been performed in bacteria, in Escherichia coli. It is unclear if these IS elements will also work in mammalian, mouse, or human cells. It is foreseeable that they might, but we should wait to see these results before getting too excited about an editing system based on mobile IS elements that promises to address the shortcomings of CRISPR systems in reconstructing severe chromosomal alterations frequently causing congenital diseases.
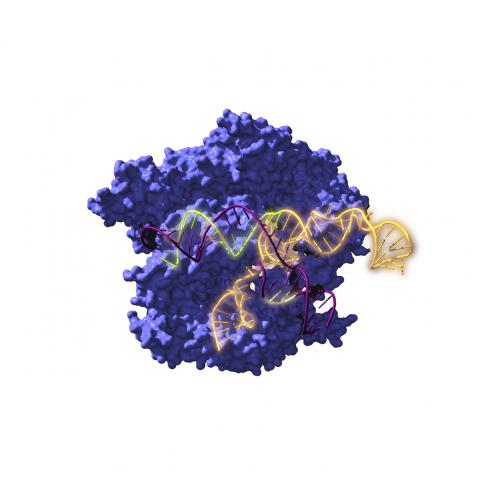
The second of Hsu's articles describes the ultrastructure of this IS system along with the bridge RNA and the DNA target and donor DNA at different points in the enzymatic reaction. These data allow for the reconstruction of all process steps and the deduction of the mechanism by which these small IS family mobile elements can promote the insertion, deletion, or inversion of DNA fragments with high (albeit variable) efficiency and cleanly, without leaving a trace, and without dragging or duplicating adjacent sequences.
DNA transposons or mobile genetic elements were discovered in the 1940s by Barbara McClintock in the maize genome and later found in practically all living organisms' genomes. McClintock had to wait nearly 40 years to receive, solo, the deserved Nobel Prize in Physiology or Medicine in 1983, recognizing the value and impact of her discovery. The proteins that mediate transposon jumps are transposases, related to CRISPR system Cas nucleases and these small IS element recombinases. I can almost sense Barbara McClintock’s smile in the lines of Hsu’s articles, reminding us once again that transposons have been and continue to be engines of change and evolution, providing us with new systems to increasingly modify and manipulate any living being’s genome with greater versatility and precision.
Hiraizumi et al.
- Research article
- Peer reviewed
Durrant et al.
- Research article
- Peer reviewed