Harmful mutations in human mitochondrial DNA corrected through gene editing
A team from the Netherlands has successfully edited pathogenic mutations in mitochondrial DNA in human cells, changes in DNA that cause disease, according to research published in PLoS Biology. The authors used a genetic tool known as a base editor. Until now, techniques derived from CRISPR have made it possible to correct mutations in nuclear DNA, and new techniques are being developed that allow mitochondrial DNA to be edited.
Santiago Restrepo - edición ADN mitocondria EN
Santiago Restrepo Castillo
Postdoctoral researcher at the University of Texas at Austin (USA)
The genetic material (DNA) in our cells is like a complex ‘instruction manual’ that defines our body's functions at the molecular level. Errors in this manual (DNA mutations) can lead to genetic diseases, which can be fatal. The development of technologies for the precise and efficient correction of these mutations is essential for the establishment of gene therapies. In the last decade, there have been major advances in the therapeutic correction of DNA mutations located in the nucleus of human cells, mainly using technologies derived from CRISPR, a platform that received the Nobel Prize in Chemistry in 2020. However, the genetic material stored in mitochondria has not been as easy to manipulate.
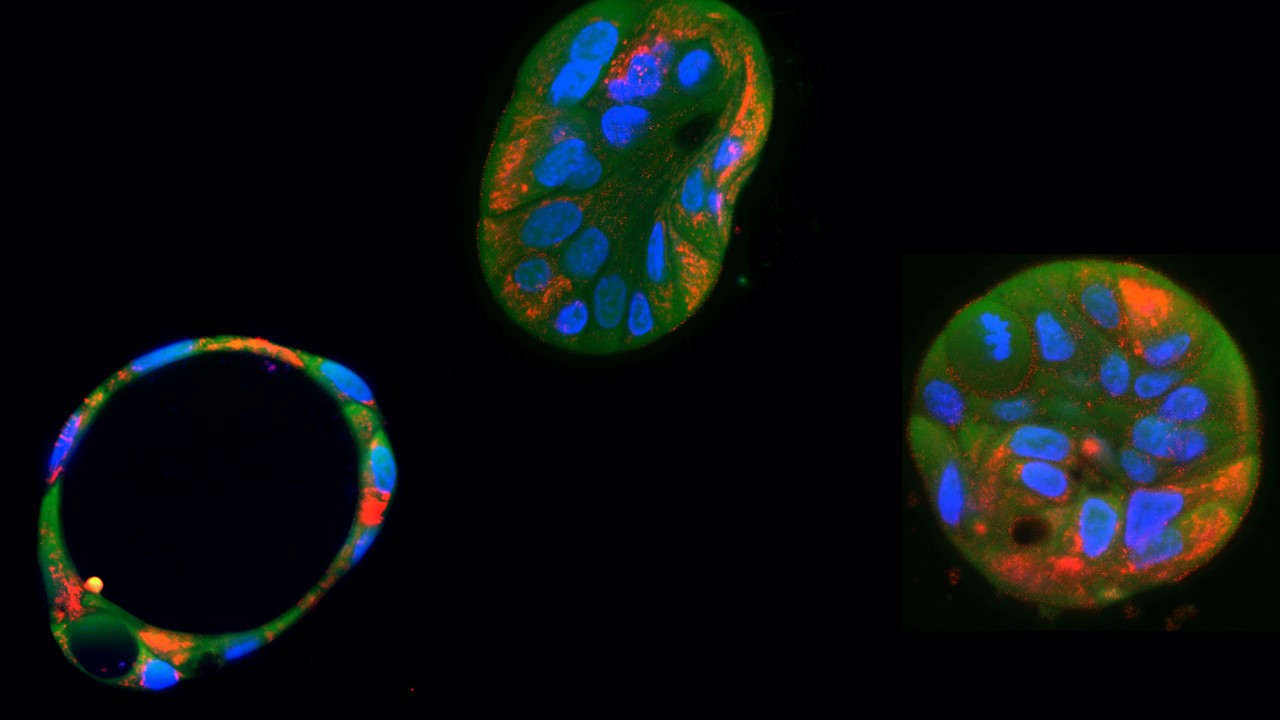
Based on previous developments of alternative technologies to CRISPR for precise editing of mitochondrial DNA, the authors of this study demonstrated for the first time the usefulness of these tools in organoids (three-dimensional cell cultures), which can provide essential information in preclinical studies. In addition, the authors corrected a mitochondrial DNA mutation in cells derived from patients, a strategy that has been explored in other studies but further demonstrates the therapeutic potential of mitochondrial DNA base editors. Finally, the authors explored the translational axis of their research by demonstrating for the first time the delivery of these editors in lipid nanoparticles, a promising strategy for the therapeutic transfer of these tools.
This study represents a promising proof of concept, which will be complemented by new advances and applications of mitochondrial DNA editors in lipid nanoparticles, particularly for the development of personalised gene therapies using organoids derived from patients with different mitochondrial mutations.
250623 ADN mitocondrial Lluis EN
Lluís Montoliu
Research professor at the National Biotechnology Centre (CNB-CSIC) and at the CIBERER-ISCIII
Microbiologist Lynn Margulis will go down in the history of biology for having proposed the endosymbiotic theory of the origin of eukaryotic cells (those that make up the organisms of yeast, fungi, plants, and animals), according to which different bacteria organized and learned to live together in symbiosis. One of these gave rise to the subcellular organelle we know today as mitochondria, which eventually specialized in producing the energy the cell needed to survive.
Mitochondria, due to their bacterial origin, have their own genome, now greatly simplified after billions of years of evolution, given that most of the genes they need to function have eventually been transferred to the cell nucleus. However, it still retains approximately 16,500 pairs of letters that encode genetic information to produce essential proteins that mitochondria need to continue generating energy. Naturally, a mutation in any of these mitochondrial genes has devastating consequences for the life of the person who inherits such a genetic alteration. That's why congenital mitochondrial diseases, which are very rare (they're called rare diseases), are so serious, most often fatal, with very complex pathological manifestations that end up affecting virtually every organ and part of the body. Since we inherit most of our mitochondria from the egg (our mitochondria are essentially maternal), congenital mitochondrial diseases are maternally inherited.
The almost all-powerful CRISPR, initially described by Francis Mojica as a defense system used by bacteria to fight off the viruses that threaten them, and transformed by Emmanuelle Charpentier and Jennifer Doudna into true gene-editing tools, cannot enter mitochondria. That's why CRISPR tools cannot be used to edit mitochondrial DNA.
David R. Liu, a researcher at the BROAD Institute in Boston, knew about the limitations of CRISPR in mitochondria, so he invented a solution, turning to the previous gene-editing system, TALENs, also derived from bacteria that infect plants. TALENs don't need RNA to identify the gene to be edited, as CRISPRs do. TALENs use a variable portion of these proteins to pair with specific DNA sequences. The N portion of TALENs is the nuclease of the system, which cuts the DNA. But if only the TALE portion is used (without the nuclease), this protein can be used to identify a mitochondrial DNA sequence. Then, simply adding a deaminase module (such as cytidine deaminase) creates mitochondrial genome base editors that can change a C to a T, a spectacular advance described by Liu in 2020. Two years later, Liu improved these mitochondrial editors by relaxing the bases that should be in positions adjacent to the one to be edited, turning them into much more versatile mitochondrial genome editing tools. Liu also developed mitochondrial base editors based on another, even earlier gene-editing system: zinc-finger proteins. Now, all this knowledge and extraordinary prior technological developments have allowed researchers from Utrecht, the Netherlands, to use Liu's evolved TALE base editors to correct, in cellular models and organoids, pathogenic mutations identified in the mitochondrial DNA of patients, pioneering the clinical use of these new mitochondrial genome editing tools for the treatment of congenital mitochondrial diseases.
This work, published in the prestigious journal PLoS Biology, reports two types of experiments, impossible to perform with CRISPR. First, these researchers used these mitochondrial base editors (technically called DdCBE) to introduce a specific mutation into the mitochondrial DNA of cells from a liver organoid (derived from human inducible pluripotent stem cells). With this, they verified the reduction in energy production and explored different levels of heteroplasmy, a condition that occurs when healthy mitochondria and mitochondria affected by the mutation coexist in cells.
Secondly, and perhaps more importantly, the researchers obtained fibroblasts from a patient suffering from one of these very serious congenital mitochondrial diseases and used the DdCBE mitochondrial base editors to correct the mutation (a T to a C at position 4291 of the mitochondrial genome), converting the C back into the correct, original T, thereby restoring the functionality of these edited mitochondria.
In this study, the researchers combine the innovative use of the most evolved versions of mitochondrial base editors with lipid nanoparticles, which allow them to target the editors to mitochondria without the need for viral vectors. These scientists also applied state-of-the-art, single-cell methods to analyze what was happening in each cell, given that each cell can contain hundreds to thousands of mitochondrial DNA molecules. They found that modification efficiency ranged from 0% to 80%, with different levels of heteroplasmy (a mixture of intact and edited mitochondria).
The latest and surprising innovation developed by these researchers is to test the introduction of DdCBE mitochondrial base editors not in protein form but in the form of modified RNAs, within lipid nanoparticles, following the design that began with the COVID-19 vaccines and continued with various therapeutic proposals that have continued to use the administration of gene editors in the form of RNA. And the results they obtained were much better than those obtained by administering the base editors in the form of DNA. Finally, the analysis of edits in other unwanted sequences of nuclear and mitochondrial DNA (off-targets) produced either non-significant results (in nuclear DNA) or some unexpected changes (in mitochondrial DNA), the significance of which correlated with the percentage of editing in the selected sequence. The possible deleterious effects of these unwanted mutations should be investigated in detail.
This work is certainly relevant, as it opens the door to treating extremely serious congenital mitochondrial diseases, which until now have been incurable, through the combined use of various cutting-edge technologies.
Gemma Marfany - crispr mitocondrias
Gemma Marfany
Professor of Genetics at the Universitat de Barcelona (UB) and head of group at CIBERER
Gene therapy for mitochondrial diseases, which are maternally inherited, is difficult because the entry of nucleic acids into the mitochondria is highly restricted. This means that these diseases, which involve mutations in mitochondrial DNA, have lagged behind in the design of therapies, as the conventional CRISPR gene editing strategy is not possible because the guide RNA cannot enter the mitochondria. The interest of this research lies in the fact that the researchers have used an alternative strategy that requires only the entry of proteins into the mitochondria. They have used base editors derived from a bacterial toxin fused to TALEN proteins that direct the editors to specific sequences of mitochondrial DNA. With this strategy, they have managed to generate a model of mitochondrial disease in liver organoids and correct, to a certain extent, fibroblasts from patients affected by a mitochondrial disease. It is also interesting that they have found that the delivery of the necessary components for base editing is more efficient if this machinery is transferred using modified RNA wrapped in lipid particles, in a manner very similar to how COVID-19 RNA vaccines were delivered.
Indi P. Joore et al.
- Research article
- Peer reviewed
- Experimental study


